Bacteria, archaea, and viruses are all around us in natural and built environments. Microbes have the ability to make us sick (pathogens) , but also have the ability to be systematically harnessed for a productive output (bio-manufacturing and wastewater treatment). We are interested in the study of complex microbial communities and how they function together, how genes transfer within them, and how we can manipulate them. We use culture and molecular methods to understand these communities and how we can leverage them.
Environmental Dissemination of Antibiotic Resistance
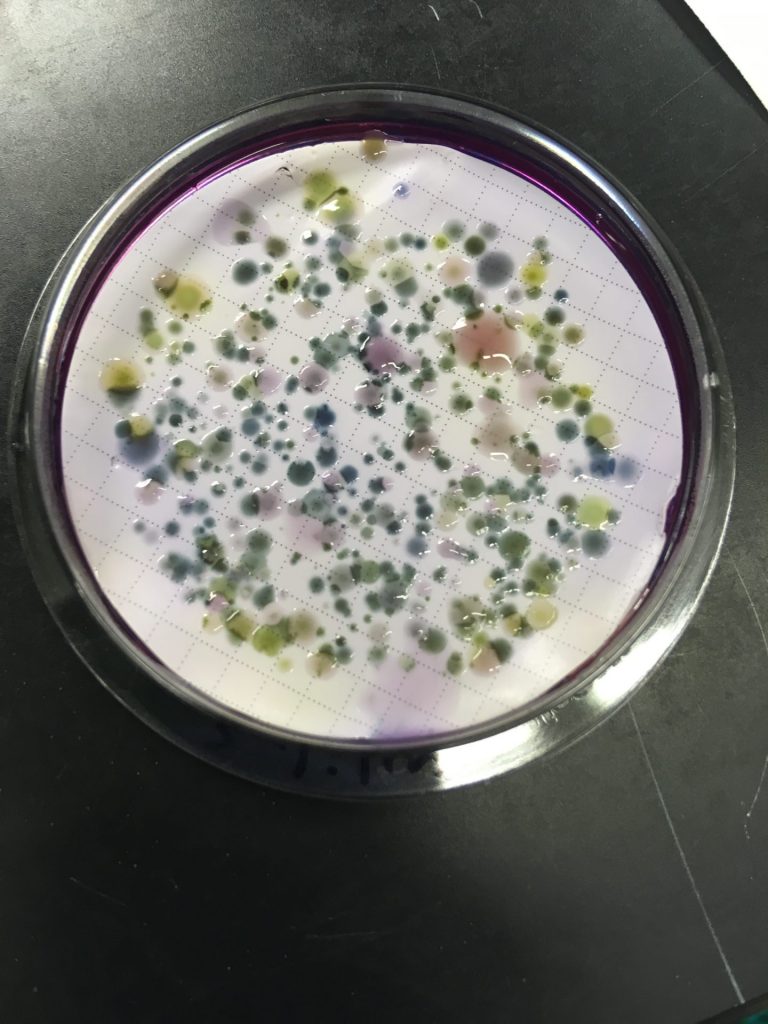
Antibiotic resistance is a known public health threat that is only growing. However, we still do not understand where resistant bacteria evolve, how to inhibit them specifically, and where exposure routes occur. Our work focuses on optimizing wastewater treatment to ensure that antibiotic resistant bacteria are removed by wastewater treatment so that they cannot impact downstream communities.
Wastewater Surveillance

Tracking pathogens in wastewater systems can improve our understanding of community health. Our group focuses on establishing methods to generate comparable wastewater based surveillance data as well as examines factors that cause for wastewater surveillance to fail by providing inaccurate results.
Disrupted Systems

Natural disasters like hurricanes and floods have the ability to allow for pathogen infiltration and contamination in drinking waters. Our work aims to understand what makes systems vulnerable to pathogen infiltration as well as works on improving methods to detect pathogens in water systems.
Bioinformatic Tools
For the most part, our group uses pre-existing tools to conduct our metagenomic analyses. Sometimes there are no pre-existing tools for the things we want to analyze. In these cases we develop our own solutions.
Data Reuse and Comparability
A key tenant of science is that we are collectively learning from each other and that we can reproduce what others have done. In order to facilitate this learning, we are committed to generating robust data and metadata for all of our works. We work with the National Microbiome Data Collaborative and the International Microbiome and Multi’omics Standards Alliance to help in these goals. We maintain an active Github Repo for all of our publications (https://github.com/ikeenum/KeenumResearchGroup).
Interested in joining our group?
Our group manual can be found here: Group Manual. Please read this over and mention it in an email to Dr. Keenum (imkeenum.at.mtu.edu)